Q Location On A Chromosome
Description
Humans normally have 46 chromosomes in each cell, divided into 23 pairs. Two copies of chromosome 6, one copy inherited from each parent, form one of the pairs. Chromosome 6 spans about 171 million DNA building blocks (base pairs) and represents between 5.5 and 6 percent of the total DNA in cells.

Identifying genes on each chromosome is an active area of genetic research. Because researchers use different approaches to predict the number of genes on each chromosome, the estimated number of genes varies. Chromosome 6 likely contains 1,000 to 1,100 genes that provide instructions for making proteins. These proteins perform a variety of different roles in the body.
Changes to chromosome 6 may include deletions or duplications of genetic material in the short (p) or long (q) arm of the chromosome in each cell, or a circular structure called ring chromosome 6. Ring chromosomes occur when a chromosome breaks in two places and the ends of the chromosome arms fuse together to form a circular structure. The constricted region of linear chromosomes is known as the centromere. Although this constriction is called the centromere, it usually is not located exactly in the center of the chromosome and, in some cases, is located almost at the chromosome's end. The regions on either side of the centromere are referred to as the chromosome's arms.
- Before this happens, each chromosome is duplicated, and both copies are joined by a centromere, resulting either in an X-shaped structure (pictured above), if the centromere is located equatorially, or a two-arm structure, if the centromere is located distally.
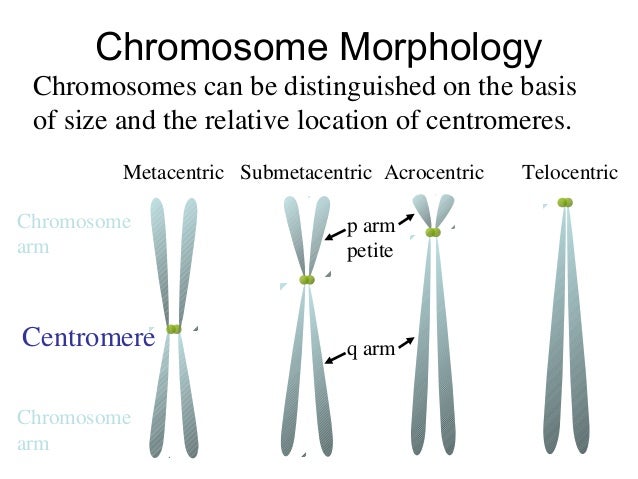
- The centromere is the connection point of the duplicated chromosome, while telomeres are the endpoints. The short arm of the chromosome is termed “p” and the long arm of the chromosome is termed “q.” If we take these two chromosome arms into consideration, there are three types of chromosome morphology.
Health Conditions Related to Chromosomal Changes
The following chromosomal conditions are associated with changes in the structure or number of copies of chromosome 6.
6q24-related transient neonatal diabetes mellitus
6q24-related transient neonatal diabetes mellitus, a type of diabetes that occurs in infants, is caused by the overactivity (overexpression) of certain genes in a region of the long (q) arm of chromosome 6 called 6q24. People inherit two copies of their genes, one from their mother and one from their father. Usually both copies of each gene are active, or 'turned on,' in cells. In some cases, however, only one of the two copies is normally turned on. Which copy is active depends on the parent of origin: some genes are normally active only when they are inherited from a person's father; others are active only when inherited from a person's mother. This phenomenon is known as genomic imprinting.
The 6q24 region includes paternally expressed imprinted genes, which means that normally only the copy of each gene that comes from the father is active. The copy of each gene that comes from the mother is inactivated (silenced) by a mechanism called methylation.
There are three ways that overexpression of paternally expressed imprinted genes in the 6q24 region can occur. About 40 percent of cases of 6q24-related transient neonatal diabetes mellitus are caused by a genetic change known as paternal uniparental disomy (UPD) of chromosome 6. In paternal UPD, people inherit both copies of the affected chromosome from their father instead of one copy from each parent. Paternal UPD causes people to have two active copies of paternally expressed imprinted genes, rather than one active copy from the father and one inactive copy from the mother.
Another 40 percent of cases of 6q24-related transient neonatal diabetes mellitus occur when the copy of chromosome 6 that comes from the father has a duplication of genetic material including the paternally expressed imprinted genes in the 6q24 region.
The third mechanism by which overexpression of genes in the 6q24 region can occur is by impaired silencing of the maternal copy of the genes (maternal hypomethylation). Approximately 20 percent of cases of 6q24-related transient neonatal diabetes mellitus are caused by maternal hypomethylation. Some people with this disorder have a genetic change in the maternal copy of the 6q24 region that prevents genes in that region from being silenced. Other affected individuals have a more generalized impairment of gene silencing involving many imprinted regions, called hypomethylation of imprinted loci (HIL). Because HIL can cause overexpression of many genes, this mechanism may account for the additional health problems that occur in some people with 6q24-related transient neonatal diabetes mellitus.
It is not well understood how overexpression of genes in the 6q24 region causes 6q24-related transient neonatal diabetes mellitus and why the condition improves after infancy. This form of diabetes is characterized by high blood sugar levels (hyperglycemia) resulting from a shortage of the hormone insulin. Insulin controls how much glucose (a type of sugar) is passed from the blood into cells for conversion to energy.
The protein produced from one gene in the 6q24 region may help control insulin secretion by beta cells in the pancreas. In addition, overexpression of this protein has been shown to stop the cycle of cell division and lead to the self-destruction of cells (apoptosis). Researchers suggest that overexpression of this gene may reduce the number of insulin-secreting beta cells or impair their function in affected individuals.
Lack of sufficient insulin results in the signs and symptoms of diabetes mellitus. In individuals with 6q24-related transient neonatal diabetes mellitus, these signs and symptoms are most likely to occur during times of physiologic stress, including the rapid growth of infancy, childhood illnesses, and pregnancy. Because insulin acts as a growth promoter during early development, a shortage of this hormone may account for the slow growth before birth (intrauterine growth retardation) seen in 6q24-related transient neonatal diabetes mellitus.
More About This Health ConditionOther chromosomal conditions
Other changes in the number or structure of chromosome 6 can have a variety of effects, including delayed growth and development, intellectual disability, distinctive facial features, birth defects, and other health problems. Changes to chromosome 6 may include deletions or duplications of genetic material in the short (p) or long (q) arm of the chromosome in each cell, or a circular structure called ring chromosome 6. Ring chromosomes occur when a chromosome breaks in two places and the ends of the chromosome arms fuse together to form a circular structure.
Cancers
Duplications of genetic material in the short (p) arm of chromosome 6 have been associated with the growth and spread of several types of cancer. These duplications are somatic, which means they are acquired during a person's lifetime and are present only in certain cells. Researchers believe that some of the genes in the duplicated region on chromosome 6p are oncogenes. Oncogenes play roles in several critical cell functions, including cell division, the maturation of cells to carry out specific functions (cell differentiation), and the self-destruction of cells (apoptosis). When mutated, oncogenes have the potential to cause normal cells to become cancerous. The presence of extra copies of the oncogenes may allow cells to grow and divide in an uncontrolled way, leading to the progression and spread of cancer.
Additional Information & Resources
Additional NIH Resources
- National Human Genome Research Institute: Chromosome Abnormalities
References
- Andrieux J, Devisme L, Valat AS, Robert Y, Frnka C, Savary JB. Prenataldiagnosis of ring chromosome 6 in a fetus with cerebellar hypoplasia and partial agenesis of corpus callosum: case report and review of the literature. Eur J Med Genet. 2005 Apr-Jun;48(2):199-206. Epub 2005 Feb 12. Review. Citation on PubMed
- Diatloff-Zito C, Nicole A, Marcelin G, Labit H, Marquis E, Bellanné-Chantelot C, Robert JJ. Genetic and epigenetic defects at the 6q24 imprinted locus in acohort of 13 patients with transient neonatal diabetes: new hypothesis raised by the finding of a unique case with hemizygotic deletion in the critical region. J Med Genet. 2007 Jan;44(1):31-7. Epub 2006 Sep 13. Citation on PubMed or Free article on PubMed Central
- Docherty LE, Poole RL, Mattocks CJ, Lehmann A, Temple IK, Mackay DJ. Furtherrefinement of the critical minimal genetic region for the imprinting disorder6q24 transient neonatal diabetes. Diabetologia. 2010 Nov;53(11):2347-51. doi:10.1007/s00125-010-1853-2. Epub 2010 Jul 30. Citation on PubMed
- Gilbert F. Chromosome 6. Genet Test. 2002 Winter;6(4):341-58. Citation on PubMed
- Lin RJ, Cherry AM, Chen KC, Lyons M, Hoyme HE, Hudgins L. Terminal deletion of6p results in a recognizable phenotype. Am J Med Genet A. 2005 Jul15;136(2):162-8. Review. Citation on PubMed
- Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC,Horton R, Hunt SE, Scott CE, Gilbert JG, Clamp ME, Bethel G, Milne S, AinscoughR, Almeida JP, Ambrose KD, Andrews TD, Ashwell RI, Babbage AK, Bagguley CL,Bailey J, Banerjee R, Barker DJ, Barlow KF, Bates K, Beare DM, Beasley H, BeasleyO, Bird CP, Blakey S, Bray-Allen S, Brook J, Brown AJ, Brown JY, Burford DC,Burrill W, Burton J, Carder C, Carter NP, Chapman JC, Clark SY, Clark G, Clee CM,Clegg S, Cobley V, Collier RE, Collins JE, Colman LK, Corby NR, Coville GJ,Culley KM, Dhami P, Davies J, Dunn M, Earthrowl ME, Ellington AE, Evans KA,Faulkner L, Francis MD, Frankish A, Frankland J, French L, Garner P, Garnett J,Ghori MJ, Gilby LM, Gillson CJ, Glithero RJ, Grafham DV, Grant M, Gribble S,Griffiths C, Griffiths M, Hall R, Halls KS, Hammond S, Harley JL, Hart EA, Heath PD, Heathcott R, Holmes SJ, Howden PJ, Howe KL, Howell GR, Huckle E, Humphray SJ,Humphries MD, Hunt AR, Johnson CM, Joy AA, Kay M, Keenan SJ, Kimberley AM, KingA, Laird GK, Langford C, Lawlor S, Leongamornlert DA, Leversha M, Lloyd CR, LloydDM, Loveland JE, Lovell J, Martin S, Mashreghi-Mohammadi M, Maslen GL, MatthewsL, McCann OT, McLaren SJ, McLay K, McMurray A, Moore MJ, Mullikin JC, Niblett D, Nickerson T, Novik KL, Oliver K, Overton-Larty EK, Parker A, Patel R, Pearce AV, Peck AI, Phillimore B, Phillips S, Plumb RW, Porter KM, Ramsey Y, Ranby SA, Rice CM, Ross MT, Searle SM, Sehra HK, Sheridan E, Skuce CD, Smith S, Smith M,Spraggon L, Squares SL, Steward CA, Sycamore N, Tamlyn-Hall G, Tester J, Theaker AJ, Thomas DW, Thorpe A, Tracey A, Tromans A, Tubby B, Wall M, Wallis JM, WestAP, White SS, Whitehead SL, Whittaker H, Wild A, Willey DJ, Wilmer TE, Wood JM,Wray PW, Wyatt JC, Young L, Younger RM, Bentley DR, Coulson A, Durbin R, Hubbard T, Sulston JE, Dunham I, Rogers J, Beck S. The DNA sequence and analysis of humanchromosome 6. Nature. 2003 Oct 23;425(6960):805-11. Citation on PubMed
- Pivnick EK, Qumsiyeh MB, Tharapel AT, Summitt JB, Wilroy RS. Partialduplication of the long arm of chromosome 6: a clinically recognisable syndrome. J Med Genet. 1990 Aug;27(8):523-6. Review. Citation on PubMed or Free article on PubMed Central
- Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification andcancer progression. J Clin Pathol. 2007 Jan;60(1):1-7. Epub 2006 Jun 21. Review. Citation on PubMed or Free article on PubMed Central
- Temple IK, Mackay DJG. Diabetes Mellitus, 6q24-Related Transient Neonatal.2005 Oct 10 [updated 2018 Sep 13]. In: Adam MP, Ardinger HH, Pagon RA, WallaceSE, Bean LJH, Mirzaa G, Amemiya A, editors. GeneReviews® [Internet]. Seattle(WA): University of Washington, Seattle; 1993-2021. Available fromhttp://www.ncbi.nlm.nih.gov/books/NBK1534/ Citation on PubMed
- Temple IK, Shield JP. 6q24 transient neonatal diabetes. Rev Endocr MetabDisord. 2010 Sep;11(3):199-204. doi: 10.1007/s11154-010-9150-4. Review. Citation on PubMed
- Urban M, Bommer C, Tennstedt C, Lehmann K, Thiel G, Wegner RD, Bollmann R,Becker R, Schulzke I, Körner H. Ring chromosome 6 in three fetuses: case reports,literature review, and implications for prenatal diagnosis. Am J Med Genet. 2002 Mar 1;108(2):97-104. Review. Citation on PubMed
- Zherebtsov MM, Klein RT, Aviv H, Toruner GA, Hanna NN, Brooks SS. Furtherdelineation of interstitial chromosome 6 deletion syndrome and review of theliterature. Clin Dysmorphol. 2007 Jul;16(3):135-40. Review. Citation on PubMed
A chromosome is a packaged unit of DNA and associated proteins.[1] The chromosome carries portions of the hereditary information of an organism. The DNA is tightly coiled many times around proteins called histones that support the chromosome structure.[2] The bound DNA units are called nucleosomes. Different kinds of organisms have different numbers of chromosomes. Humans have 23 pairs of chromosomes, 46 in all: 44 autosomes and two sex chromosomes. Each parent contributes one chromosome to each pair, so children get half of their chromosomes from their mothers and half from their fathers.
History
The word 'chromosome' means 'colored body' and comes from the Greek χρώμα or 'chroma' for 'color' and σομε or 'some' for 'body'. By 1900 scientists knew that cells were the building blocks of living things. All living things start as a single fertilized cell which keeps dividing. Scientists identified tiny threads in the nucleus of the cells. Because they could stain the threads with colored dyes to study them under a microscope, they called the threads chromosomes, from the Greek words for colored bodies. They could see that chromosomes came in pairs, and that human cells all contained 23 matching pairs. American biologist Walter Sutton knew Mendel's principles of genetics work on peas, and suggested that chromosomes held the secret of inheritance.
Another American biologist, Thomas Hunt Morgan, developed the idea that chromosomes were made up of linked groups of factors called genes. He experimented with red-eyed fruit flies and found that sometimes a white-eyed fly appeared. When he mated them, he found that as he expected there were three red-eyed flies to every white-eyed fly, but that all the white-eyed flies were male. He concluded that the gene for white eyes must be on a chromosome that was related to being male. Later workers found that this is why some hereditary diseases such as hemophilia and muscular dystrophy only show in males, though women can carry the gene for the disease without showing it.
In 1991 a project called the Human Genome Project began to use computers to map the three billion base pairs which make up the 46 human chromosomes.
Q Location On A Chromosome Gene
Physical Characteristics of Chromosomes
Most chromosomes have the rough shape of an I when they condense prior to replication and are as bits of long invisible string when unwound. The familiar X shape actually refers to 2 identical chromosomes referred to as sister chromatids. Each chromosome has a constriction point called the centromere, which divides the chromosome into two sections, or “arms.” The short arm of the chromosome is labeled the “p arm” for 'petite'. The long arm of the chromosome is labeled the “q arm” for 'not petite'. The location of the centromere on each chromosome gives the chromosome its characteristic shape, and can be used to help describe the location of specific genes.[3]
The chromosomal DNA molecule contains three specific nucleotide sequences which are required for replication: a DNA replication origin; the centromere to attach the DNA to the mitotic spindle.; a telomere located at each end of the linear chromosome.[4] There is also a telomere region within the human chromosome two, as well as a non-functional second centromere.[5] This indicates a historical chromosome fusion event.
Number of Chromosomes
As noted above, the chromosome number varies in different species. In humans there are 46 chromosomes, or 23 pairs of chromosomes (diploid), in every cell except the mature egg and sperm which have a set of 23 chromosomes (haploid).
Cell Division
Chromosomes are visible only during cell division, when the DNA is super coiled and condensed to facilitate distribution into daughter cells. Cell division in somatic cells (mitosis) results in the creation of daughter cells with the same number of chromosomes as the original cell, a total of 46 chromosomes in a human. Cell division in the germ cells, eggs and sperm (meiosis), results in the creation of daughter cells with half the number of chromosomes as the original cell (haploid cells). This reduction in the number of chromosomes is important so that the original number of chromosomes is restored following fertilization of the egg by the sperm.
Karyotype Analysis
The chromosome constitution of an individual, karyotype, can be analyzed following tissue culture of an appropriate sample. The most commonly used sample is blood (using the white blood cells or lymphocytes) since it is the most accessible. However, other samples are used depending upon the indication: amniotic fluid cells, to analyze the karyotype of the fetus; products of conception, to analyze the cause of a miscarriage or stillbirth; bone marrow cells, to diagnose the presence or type of leukemia; and skin, to determine the presence of another cell line (mosaicism).
Q Location On A Chromosome Deletion
Cell division is arrested during metaphase, when the chromosome material is condensed. Following hypotonic treatment and fixation, the cells are dropped on a slide and then stained. At least 20 metaphase spreads are analyzed and 2 or 3 metaphase spreads are photographed. The chromosomes are arranged from largest to smallest to create a karyotype.
Chromosomes vary in size and in shape. The pairs of autosomal chromosomes are arranged in a karyotype from the biggest to the smallest. The sex chromosomes are placed to the right of the smallest autosomes. Chromosomes vary in shape depending upon the position of the centromere, the structure that holds the two arms of the chromosomes together. If the centromere is in the middle, the chromosome is metacentric and the chromosome arms are equal in size. If the centromere is off center, the chromosome is submetacentric with a short arm labeled p (for petite) and a long arm labeled q (the next letter after p). If the centromere is close to the end, the chromosome is acrocentric and the very short arm consists of a stalk and a knob (satellite). Based upon size and shape, human chromosomes are divided into eight groups: A (1 to 3), B (4 and 5), C (6 to 12), D (13 to 15), E (16 to 18), F (19 and 20), G (21 and 22) and the sex chromosomes, XX in females and XY in males.
Chromosomes are further identified by banding patterns created by specialized staining procedures to produce G bands, R bands, C bands, etc. The choice of staining procedure depends upon the information that is desired. Most commonly, chromosomes are stained with trypsin-Giemsa to produce the G-banded pattern. The banding pattern is determined by the degree of chromatin condensation and the specific DNA-protein present at the different sites on the chromosome. These banding patterns are distinct and consistent for each chromosome. Thus, one can reliably identify the chromosome pairs (e.g., although of the same size and shape, the six acrocentric chromosomes in the D group can be differentiated into pairs 13, 14 or 15 based upon banding patterns). The bands are individually numbered (e.g., 11q23 refers to band #23 on the long arm q of chromosome #11). :[6]
References
- ↑http://learn.genetics.utah.edu/units/disorders/karyotype/whatarechrom.cfm
- ↑http://ghr.nlm.nih.gov/handbook/basics/chromosome
- ↑http://www.rothamsted.ac.uk/notebook/courses/guide/chromo.htm
- ↑http://ghr.nlm.nih.gov/ghr/chromosomes
- ↑http://www.pnas.org/content/88/20/9051.abstract
- ↑http://www.vivo.colostate.edu/hbooks/genetics/medgen/chromo/cytotech.html